Frontiers |
您所在的位置:网站首页 › certain types of barcodes › Frontiers |
Frontiers
ORIGINAL RESEARCH article
Front. Plant Sci., 09 May 2023Sec. Plant Systematics and Evolution
Volume 14 - 2023 |
https://doi.org/10.3389/fpls.2023.1160535
Plastome evolution and phylogenomics of Trichosporeae (Gesneriaceae) with its morphological characters appraisal Yan-Fang Cui1†, Yan-Fang Cui1†,  Peng Zhou1†, Peng Zhou1†,  Kun-Li Xiang2, Kun-Li Xiang2,  Qiang Zhang3, Qiang Zhang3,  Hua Yan1, Hua Yan1,  Li-Guo Zhang1, Li-Guo Zhang1,  Bo Pan3, Bo Pan3,  Yu-Song Huang3, Yu-Song Huang3,  Zhi-You Guo4, Zhi-You Guo4,  Zhen-Yu Li2 and Zhen-Yu Li2 and  Xiao-Guo Xiang1*1Jiangxi Province Key Laboratory of Watershed Ecosystem Change and Biodiversity, Institute of Life Science and School of Life Sciences, Nanchang University, Nanchang, Jiangxi, China2State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China3Guangxi Key Laboratory of Plant Conservation and Restoration Ecology in Karst Terrain, Guangxi Institute of Botany, Guangxi Zhuang Autonomous Region and Chinese Academy of Sciences, Guilin, China4Qiannan Normal College for Nationalities, College of Biological Sciences and Agriculture, Duyun, Guizhou, China Xiao-Guo Xiang1*1Jiangxi Province Key Laboratory of Watershed Ecosystem Change and Biodiversity, Institute of Life Science and School of Life Sciences, Nanchang University, Nanchang, Jiangxi, China2State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China3Guangxi Key Laboratory of Plant Conservation and Restoration Ecology in Karst Terrain, Guangxi Institute of Botany, Guangxi Zhuang Autonomous Region and Chinese Academy of Sciences, Guilin, China4Qiannan Normal College for Nationalities, College of Biological Sciences and Agriculture, Duyun, Guizhou, ChinaTrichosporeae is the largest and most taxonomically difficult tribe of Gesneriaceae due to its diverse morphology. Previous studies have not clarified the phylogenetic relationships within this tribe on several DNA markers, including the generic relationships within subtribes. Recently, plastid phylogenomics have been successfully employed to resolve the phylogenetic relationships at different taxonomic levels. In this study, plastid phylogenomics were used to explore the relationships within Trichosporeae. Eleven plastomes of Hemiboea were newly reported. Comparative analyses, phylogeny and morphological character evolution within Trichosporeae were conducted on 79 species representing seven subtribes. The Hemiboea plastomes range from 152,742 bp to 153,695 bp in length. Within Trichosporeae, the sampled plastomes range from 152,196 bp to 156,614 bp and GC content from 37.2% to 37.8%. A total of 121–133 genes were annotated in each species, including 80–91 protein-coding genes, 34–37 tRNA genes, and 8 rRNA genes. The contraction and expansion of IR borders were not detected, and gene rearrangements and inversions did not occur. The 13 hypervariable regions were proposed as the potential molecular markers for species identification. A total of 24,299 SNPs and 3,378 indels were inferred, and most of the SNPs were functionally missense and silent variations. There were 1968 SSRs, 2055 tandem repeats and 2802 dispersed repeats. The RSCU and ENC values indicated that the codon usage pattern was conserved in Trichosporeae. Both the phylogenetic frameworks based on the whole plastome and 80 CDSs were basically concordant. The sister relationships between Loxocarpinae and Didymocarpinae were confirmed, and Oreocharis was a sister group of Hemiboea with high support. The morphological characters showed a complex evolutionary pattern of Trichosporeae. Our findings may contribute to future research on genetic diversity, morphological evolutionary patterns, and conservation of the tribe Trichosporeae. 1 IntroductionGesneriaceae comprises ca. 150 genera and 3000 species and mainly distributes in the tropics and subtropics of the world (Li and Wang, 2005; Möller et al., 2016). These plants are usually perennial herbs with gorgeous flowers and various leaves, which have high ornamental value and great potential for garden and horticultural applications (Li and Wang, 2005). For example, Sinningia speciosa Benth. & Hook., Episcia cupreata (Hook.) Hanst. and Streptocarpus ionanthus (H. Wendl.) Christenh. have become famous ornamental flowers. Furthermore, many Gesneriaceae species, such as Corallodiscus lanuginosus (Wall. ex R. Br.) B.L. Burtt, Dorcoceras hygrometricum Bunge, Hemiboea subcapitata Clarke, Lysionotus pauciflorus Maxim. and Primulina eburnea (Hance) Yin Z. Wang, have been used as folk medicines in China for a long time (Li and Wang, 2005). As the largest tribe of Gesneriaceae, Trichosporeae includes 10 subtribes and distributes in tropical and subtropical Asia, Europe and Africa. Its morphological characters are diverse, and traditional classifications based on morphology have identified genera that belong to different alliances and geographical groups. This makes it the most taxonomically difficult tribe (e.g., Weber et al., 2013; Möller et al., 2016). Phylogenetically, the tribe is subdivided into distinct clades, but their relationships are not fully resolved and not consistently well supported (e.g., Wang et al., 2010; Möller et al., 2011; Li et al., 2022). Based on 16 plastid, 9 nuclear and 1 mitochondrial DNA makers, Roalson and Roberts (2016) showed that Streptocarpinae and Loxocarpinae were sister groups, while Li et al. (2022), based on 11 chloroplast regions and nuclear ITS, indicated that Loxocarpinae was a sister group to Didymocarpinae. Besides, the phylogenetic positions of some genera within Didymocarpinae were also uncertain. For example, both Roalson and Roberts (2016) and Li et al. (2022) indicated that Hemiboea was close to Lysionotus with weak supporting values, while Hsieh et al. (2022) supported that Oreocharis was a sister group of Hemiboea based on plastomes with one IR excluded. Möller et al. (2011) proposed to employ more molecular markers, especially conserved plastome sequences, to improve the phylogenetic relationships within Trichosporeae. Chen et al. (2020) also intensively suggested adopting more DNA sequences to resolve the phylogeny of Gesneriaceae. Therefore, developing more effective molecular markers is crucial for the conservation and utilization of Trichosporeae as well as Gesneriaceae. With the advancement of high-throughput sequencing technologies, more and more plastomes are being successfully sequenced and annotated (Dobrogojski et al., 2020). Most angiosperms plastomes are circular with four regions: a large single copy (LSC) region, a small single copy (SSC) region and two copies of inverted repeats (IRa/b), with a length ranging from 120–160 kb with 30–40% GC content (Mower and Vickrey, 2018). The high conservatism and slow evolution rate of plastomes make them suitable for inferring phylogenetic relationships (e.g., Lu et al., 2016; Chi et al., 2018; Lee et al., 2019). Rosaceae is one of the taxonomically difficult lineages with hybridization and rapid radiation, and the phylogenetic relationships among subfamilies, tribes and genera have been successfully resolved by employing plastid phylogenomics (Zhang et al., 2017). Wei et al. (2021) clarified the phylogenetic relationships within Euphorbia based on plastomes, which have highly homogenous morphological characters, and revealed that it may have undergone complex plastome evolution. Overall, plastome data has a high power to explore the phylogenetic relationships of different taxonomic levels. In this study, we newly sequenced, assembled, and annotated plastomes of 11 Hemiboea species, and combined them with 68 previously reported plastomes of Trichosporeae, we conducted comparative analyses, phylogenetic reconstruction and morphological characters appraisal within Trichosporeae. Our objectives are: (1) to investigate general plastome features and sequence divergence in Trichosporeae; (2) to identify the most variable regions as potential DNA barcodes for future species identification within Trichosporeae; (3) to explore the phylogenetic relationships within this tribe; (4) to assess the morphological evolution of Trichosporeae. Our study provides abundant information for the phylogeny, biogeography, and conservation of Trichosporeae. 2 Materials and methods2.1 Sampling, sequencing, assembly, and annotationIn this study, eleven new plastomes of Hemiboea species were obtained. Sixty-eight published plastomes of Trichosporeae were downloaded from GenBank and updated the annotations (one species of Actinostephanus, two species of Beccarinda, one species of Boeica, one species of Corallodiscus, one species of Dorcoceras, one species of Haberlea, one species of Henckelia, one species of Leptoboea, one species of Litostigma, one species of Lysionotus, five species of Oreocharis, 12 species of Paraboea, four species of Petrocodon, 28 species of Primulina, two species of Rhynchotechum, one species and five varieties of Streptocarpus), representing seven subtribes (Corallodiscinae, Didymocarpinae, Leptobaeinae, Litostigminae, Loxocarpinae, Ramondinae and Streptocarpinae). Additionally, two species of Gesnerioideae (Achimenes cettoana H.E. Moore and Achimenes erecta (Lam.) H.P. Fuchs) were selected as outgroups. The detailed information of samples was listed in Table S1. Total DNA was extracted from silica gel-dried leaves using the modified CTAB method (Doyle and Doyle, 1987). Library construction was performed with the NEB Next® Ultra DNA Library Prep Kit (NEB, USA) following the manuals. Libraries for paired-end 150 bp sequencing were conducted using an Illumina HiSeq 2000 platform to generate approximately 4 Gb of data for each sample. The library preparation and sequencing were finished at the Kunming Institute of Botany, Chinese Academy of Sciences (Yunnan, China). The quality of raw sequence reads was assessed in FastQC v0.11.9 (Brown et al., 2017) and the adapters and low-quality reads were filtered in Trimmomatic v0.39 (Bolger et al., 2014). Then, the clean reads were assembled using GetOrganelle v1.7.3.2 with default parameters (Jin et al., 2020). The assembled genomes were checked and visualized in Bandage v0.7.1 (Wick et al., 2015). Finally, the plastomes were annotated and manually checked in Geneious v9.05 (Kearse et al., 2012) with Hemiboea ovalifolia (W.T. Wang) A. Weber & Mich. Möller (NC_054358) as a reference. The physical map was drawn using OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html). 2.2 Structure and sequence divergence analysisThe expansion and contraction of IR boundary were visualized in IRscope v3.1 (Amiryousefi et al., 2018). Sequence alignments were performed using MAFFT v7 (Katoh and Standley, 2013) and manually adjusted in Geneious v9.05 (Kearse et al., 2012). Before the collinearity analysis, one copy of the IR region was removed. Possible rearrangements and inversions were detected using the Mauve v1.1.3 (Darling et al., 2010) plugin in Geneious v9.05 (Kearse et al., 2012). To further detect hypervariable regions within Trichosporeae, nucleotide diversity (pi) of protein-coding genes (CDSs) and non-coding regions were evaluated using DnaSP v6.12.03 (Rozas et al., 2017). Functional annotations for the nucleotide variations within Trichosporeae plastomes were performed using snpEff v5.1 (Cingolani, 2012). 2.3 Repetitive sequence analysisThree types of repeats sequences, including simple sequence repeats (SSRs), tandem repeats, and dispersed repeats, were analyzed. SSRs were detected using MISA v2.1 (Beier et al., 2017) and visualized using R packages ggpubr and ggplot2 (Wickham, 2016; Kassambara, 2023). Tandem repeats were performed using Tandem Repeats Finder v0.9 (Benson, 1999). The identification of dispersed repeats (including forward, reverse, complement, and palindromic) was analyzed in REPuter (Kurtz et al., 2001) following the parameter settings in Cauz-Santos et al. (2020). 2.4 Codon usage analysisTo quantify the degree of the codon usage bias, both the relative synonymous codon usage (RSCU) ratio and the effective number codons (ENC) for 79 Trichosporeae plastomes were estimated using CodonW v1.4.2 (Peden, 1999). The RSCU>1 indicates a preferred codon, while the RSCU0.025; Figure 3A). Among the non-coding regions, the pi values ranged from 0 (i.e., rpoC1/rpoB, trnI-GAU/trnA-UGC and ndhA/ndhH) to 0.409580 (rpl16/rps3). Eight non-coding regions (i.e., rpl16/rps3, rpl14/rpl16, petB/petD, trnC-GCA/petN, rps15/ycf1, trnfM-CAU/rps14, petD/rpoA and trnS-GGA/rps4) showed significantly high pi values (pi>0.25; Figure 3B). The aligned matrix of the 79 Trichosporeae plastomes contained 24,299 single nucleotide polymorphisms (SNPs) and 3378 insertion-deletions (indels). The majority of SNPs from coding genes were functionally missense variations, while 6448 SNPs (43.20%) and 104 SNPs (0.70%) were silent (synonymous) and nonsense variations, respectively (Table 1). FIGURE 3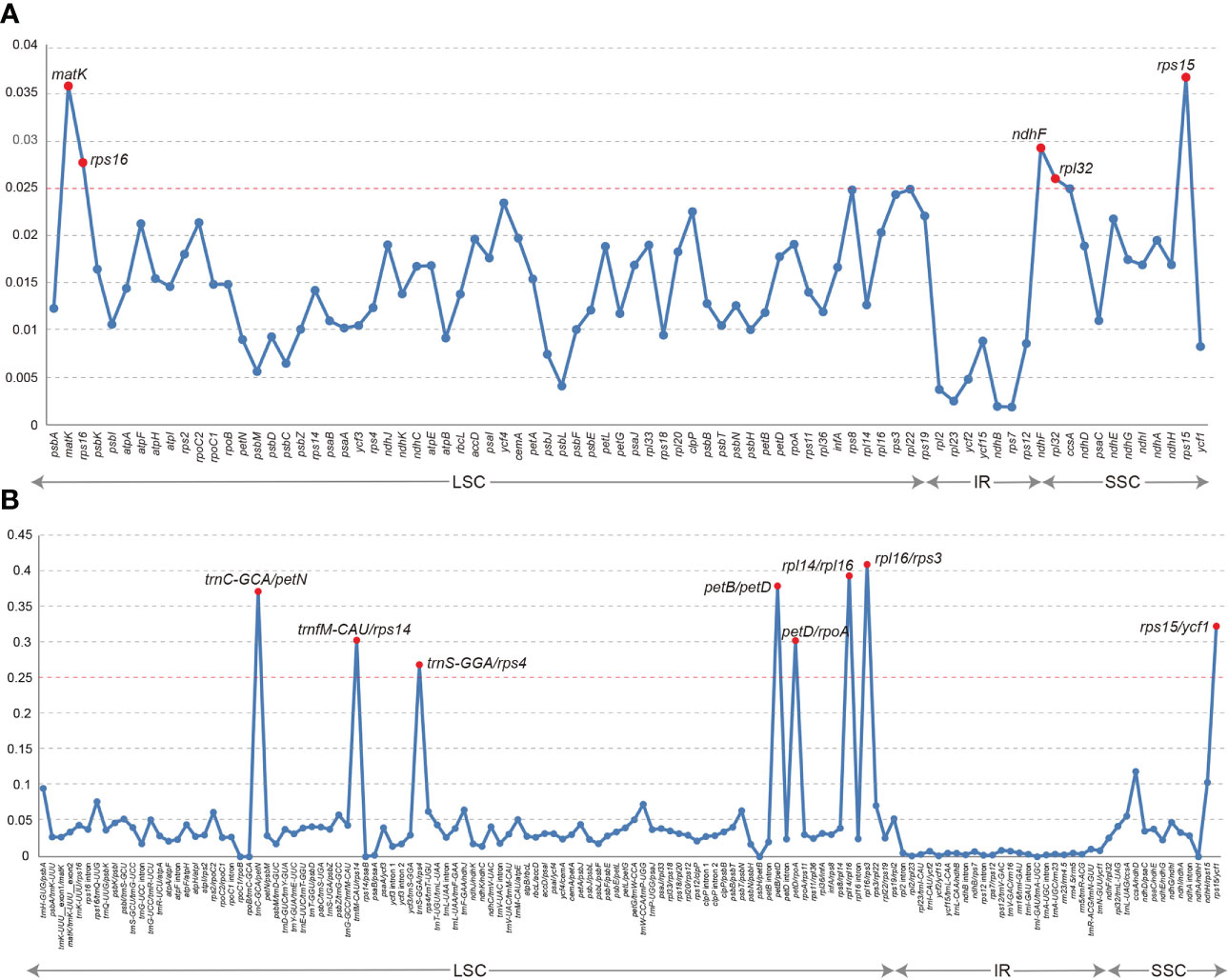 Figure 3 Comparison of nucleotide diversity (Pi) values in Trichosporeae plastomes. (A) Pi values of CDSs, (B) Pi values of non-coding regions. TABLE 1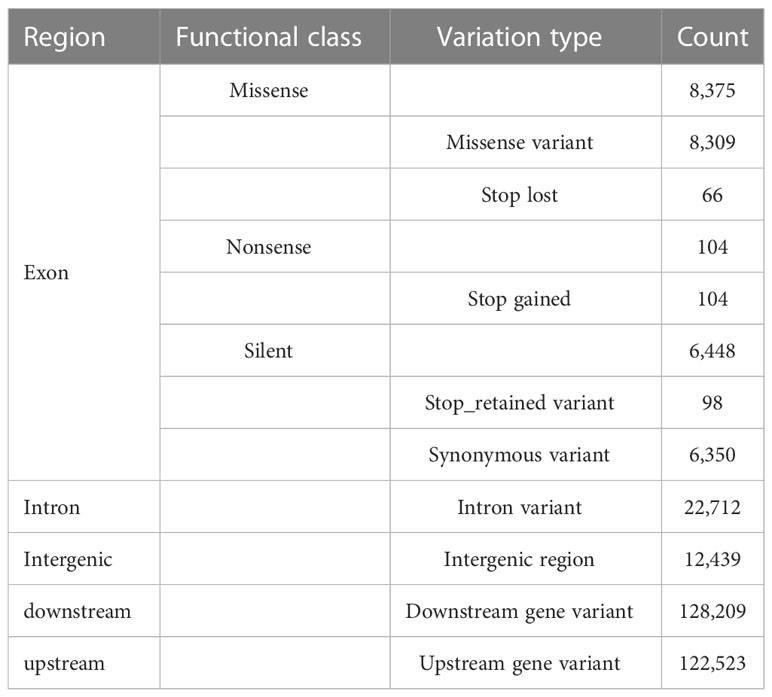 Table 1 Functional annotaitons for the single-nucleotide variants (SNVs) detected in the Trichosporeae plastomes. The results of SSRs, tandem repeats, and dispersed repeats in the 79 Trichosporeae plastomes were shown in Figure 4 and Table S4. The number of identified SSRs ranged from 11 (Dorcoceras hygrometricum) to 53 (Corallodiscus lanuginosus). A total of 1968 SSRs were detected in the 79 Trichosporeae plastomes, of which 1432 SSRs (62.60%) located in the LSC region, 280 SSRs (14.23%) in the SSC region, and 258 SSRs (13.11%) in the IR regions. Four types of SSRs (mono-nucleotide, di-nucleotide, tri-nucleotide and tetra-nucleotide) were identified, with 1771 SSRs (89.99%) being mono-nucleotide type, particularly A and T repeat motifs. In the di-nucleotide repeat type, 176 SSRs were AT/TA repeat motifs. Only one TC and AG repeat motifs were found in H. ovalifolia, Dorcoceras hygrometricum, Litostigma coriaceifolium Y. G. Wei, F. Wen & Mich. Möller, Primulina lobulata (W.T. Wang) Mich. Möller & A. Weber and Primulina xiziae F. Wen, Yue Wang & G. J. Hua, respectively. In tri-nucleotide repeat type, only TTA, ATA and ATT repeat motifs were found in Paraboea filipes (Hance) Burtt, P. peltifolia D. Fang & L. Zeng, P. clavisepala D. Fang & D. H. Qin, P. dictyoneura (Hance) Burtt, P. dolomitica Z.Y. Li, X.G. Xiang & Z.Y. Guo and Primulina fimbrisepala (Hand.-Mazz.) Yin Z. Wang. Similarly, in the tetra-nucleotide repeat type, only one ATCT repeat motif was found in H. malipoensis Y. H. Tan, H. sinovietnamica, H. subacaulis Hand.-Mazz. Petrocodon jingxiensis (Yan Liu, H. S. Gao & W. B. Xu) A. Weber & Mich. Möller, and Primulina linearifolia (W. T. Wang) Yin Z. Wang. A total of 2055 tandem repeats were detected, ranging from 13 (Petrocodon longitubus Cong R. Li & Yang Luo) to 47 (Corallodiscus lanuginosus). There were 2802 dispersed repeats in the Trichosporeae plastomes, of which 1185 (42.29%) were forward repeats, 1502 (53.60%) were palindromic repeats, 86 (3.07%) were reverse repeats, and 29 (1.03%) were complement repeats. FIGURE 4 Figure 4 Plot of each SSR repeat pattern numbers of 79 Trichosporeae plastomes with one IR excluded. The total number of SSRs in each plastome is shown in parentheses. 3.3 Codon usage biasThe codon usage bias of CDSs in Trichosporeae was shown in Figures S2, 3. There were 30 preferred codons (RSCU>1), 2 non-preferred codons (RSCU=1), and 32 less frequently used codons (RSCU |
【本文地址】